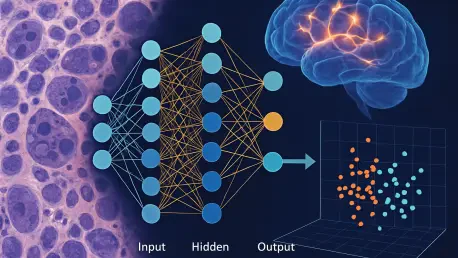
Neuroblastoma, recognized as one of the most devastating cancers affecting children, continues to challenge pediatric oncology with its alarmingly low survival rates for high-risk patients, which hover below 60%, making it a critical area of research. This aggressive tumor, frequently diagnosed in infants, accounts for roughly 15% of all childhood cancer deaths, even with interventions like surgery, chemotherapy, and stem cell therapies. Existing biomarkers, such as MYCN amplification and ALK mutations, often fall short, lacking the breadth to predict outcomes across diverse patient groups or guide tailored treatments effectively. This persistent gap in reliable prognostic tools has left clinicians struggling to improve survival odds. However, a pioneering study from the Children’s Hospital of Chongqing Medical University, recently published in Pediatric Discovery in May of this year, introduces a transformative approach. By leveraging an advanced machine learning framework, the research team analyzed RNA sequencing data from over 1,200 patients to identify novel genetic markers that could redefine how this disease is understood and managed.
Harnessing Computational Tools for Cancer Insights
Decoding Genetic Signatures
The application of machine learning in cancer research has opened new frontiers, particularly in tackling complex diseases like neuroblastoma. This study employed cutting-edge computational tools, such as an enhanced stSVM algorithm, to sift through bulk RNA sequencing data from a vast cohort of patients, identifying 528 genes associated with survival outcomes. Through further refinement using weighted gene co-expression network analysis (WGCNA), the list was narrowed to 11 critical hub genes, with RFC3 emerging as a standout marker. This process showcases the ability of machine learning to handle massive datasets and uncover hidden patterns in genetic expression that traditional methods might overlook. The focus on RFC3, linked to poorer prognosis, underscores how such technology can pinpoint specific targets for deeper investigation, potentially altering the trajectory of patient care by highlighting previously unrecognized indicators of disease severity.
Beyond the identification of these genes, the technical sophistication of this approach lies in its integration of diverse data types. The combination of bulk and single-cell RNA sequencing provides a multi-layered view of neuroblastoma’s genetic landscape, revealing not just which genes matter, but how they interact within the broader context of tumor biology. This dual-method analysis ensures a more robust understanding, as it captures both overarching trends in gene expression and the nuanced behavior of individual cell types. The result is a clearer picture of how genetic markers like RFC3 contribute to disease progression, offering a foundation for developing more precise diagnostic tools. Such advancements signal a shift in oncology toward data-driven strategies that prioritize comprehensive insights over isolated findings, setting a new standard for biomarker discovery in pediatric cancers.
Embracing a Holistic Perspective
The shift from single-marker strategies to a systems-level approach marks a significant evolution in cancer research. Historically, prognostic tools for neuroblastoma relied on isolated genetic indicators, which often failed to capture the full complexity of the disease. In contrast, this study integrates multi-omics data—spanning genomic, transcriptomic, and clinical information—to build a more complete prognostic model. This reflects a growing consensus among researchers that understanding cancer requires looking at the bigger picture, where multiple factors interplay to influence outcomes. The use of machine learning to map these interactions offers a way to detect patterns that simpler analyses might miss, providing a richer framework for assessing patient risk and tailoring interventions.
Moreover, this comprehensive methodology aligns with the broader movement in oncology toward precision medicine. By accounting for the intricate web of genetic and environmental factors, the research paves the way for customized treatment plans that address the unique profile of each patient. The emphasis on integrating diverse data sources also highlights the limitations of older approaches, which often lacked the depth to inform clinical decisions effectively. As a result, this work not only advances the understanding of neuroblastoma but also contributes to a paradigm shift in how aggressive cancers are studied and managed. The potential to apply such models across different cancer types further amplifies the impact, suggesting that this integrative approach could become a cornerstone of future research efforts.
Exploring Genetic Links to Immune and Therapeutic Outcomes
Unraveling Immune Interactions
A critical finding from this research is the connection between genetic markers like RFC3 and the immune landscape of neuroblastoma tumors. High expression of RFC3 was found to correlate with reduced activity of natural killer (NK) cells and T cell infiltration, key components of the body’s defense against cancer. This suggests that RFC3 may play a role in enabling tumors to evade immune detection, a mechanism often seen in more aggressive cases. Understanding this relationship provides vital clues about why certain patients experience worse outcomes, as diminished immune response can allow tumors to grow unchecked. Such insights could lead to strategies that boost immune activity in high-risk patients, counteracting the suppressive effects associated with specific genetic profiles.
Further exploration through single-cell RNA sequencing revealed that RFC3 expression is particularly elevated in epithelial and myeloid cells within the tumors of patients with shorter survival times. This granular view of the tumor microenvironment highlights how specific cell types contribute to disease dynamics, offering a more detailed map of where and how immune suppression occurs. These findings emphasize the importance of looking beyond broad genetic trends to understand the cellular context in which these markers operate. By identifying the specific environments where RFC3 exerts its influence, researchers can begin to design targeted interventions that address immune evasion at its source, potentially improving the effectiveness of existing therapies and opening new avenues for treatment development.
Guiding Treatment Through Genetic Clues
One of the most promising aspects of this study is its revelation of how genetic markers can predict responses to treatment. Tumors exhibiting high RFC3 expression demonstrated increased sensitivity to standard chemotherapy drugs like vincristine and cyclophosphamide, commonly used in neuroblastoma care. This correlation suggests that RFC3 could serve as a valuable tool for forecasting how individual patients might respond to specific medications, enabling clinicians to select therapies with the greatest likelihood of success. Such predictive power is a crucial step toward minimizing trial-and-error in treatment planning, reducing the burden of ineffective therapies on young patients while maximizing therapeutic impact.
This genetic-therapeutic linkage also aligns seamlessly with the push toward precision medicine in pediatric oncology. By integrating RFC3 expression data into clinical decision-making, healthcare providers can better stratify patients by risk and customize treatment regimens to match their unique biological profiles. This approach not only enhances the potential for positive outcomes but also mitigates the risk of overtreatment, which can have severe side effects in children. The ability to anticipate drug sensitivity through genetic markers represents a practical application of research findings, bridging the gap between laboratory discoveries and bedside care. As such, it underscores the transformative potential of machine learning in crafting more effective, individualized strategies for managing aggressive cancers.
Navigating Obstacles and Future Possibilities
Overcoming Barriers to Clinical Integration
While the computational framework developed in this study marks a significant advancement, translating these findings into clinical practice presents notable challenges. The machine learning model, though powerful, requires extensive experimental validation to ensure its reliability in real-world settings. Additional testing across diverse patient populations is essential to confirm that the identified biomarkers, such as RFC3, consistently predict outcomes and treatment responses. Furthermore, incorporating other data types, like proteomic or metabolomic profiles, could enhance the model’s accuracy by providing a more complete picture of tumor biology. These steps are critical to building trust in the technology among healthcare providers and ensuring that it can be seamlessly integrated into diagnostic and treatment workflows.
Another hurdle lies in the practical implementation of such advanced tools in clinical environments, where resources and expertise may vary widely. Developing user-friendly platforms that allow non-specialists to interpret complex genetic data will be key to widespread adoption. Additionally, addressing ethical considerations, such as patient data privacy and the potential for over-reliance on algorithmic predictions, must remain a priority. Despite these obstacles, the groundwork laid by this research offers a clear path forward. With continued refinement and collaboration between researchers and clinicians, the insights gained from this study could eventually become a routine part of pediatric cancer care, improving risk assessment and treatment precision for countless patients.
Broadening the Scope of Impact
Looking ahead, the implications of this research extend far beyond neuroblastoma, with the potential to influence the study of other aggressive cancers. The integrative machine learning pipeline developed by the team could be adapted to analyze genetic and molecular data from various cancer types, identifying prognostic markers and therapeutic targets in diseases with similarly complex profiles. This versatility positions the framework as a valuable tool in the broader fight against cancer, offering hope for advancements in areas where current diagnostic and treatment options remain limited. The ability to apply such a model across different contexts highlights the scalability of data-driven approaches in modern oncology.
Reflecting on the work completed, the study’s success in uncovering biomarkers like RFC3 through a sophisticated blend of bulk and single-cell RNA sequencing sets a precedent for future explorations. The recognition that a single gene can influence survival, immune response, and drug sensitivity provides a multifaceted view of cancer biology that had not been fully appreciated before. As researchers build on these discoveries in the months following the study’s release, the focus shifts to actionable next steps, such as designing clinical trials to test RFC3-targeted interventions. The ultimate aim is clear: to refine these computational insights into tangible solutions that can elevate survival rates and quality of life for children facing this formidable disease.