The ability to precisely visualize and manipulate molecular machinery inside living cells has long represented a monumental challenge in biological research, but a new fusion of artificial intelligence and protein engineering is rapidly turning this ambition into reality. The development of AI-driven pipelines for intrabody design represents a significant advancement in cell biology and medicine. This review will explore the evolution of this technology, its key features, performance metrics, and the impact it has had on various applications. The purpose of this review is to provide a thorough understanding of the technology, its current capabilities, and its potential future development.
The Challenge of Intracellular Targeting and the AI Solution
For decades, antibodies have served as indispensable tools for detecting and targeting molecules, yet their utility has been largely confined to the extracellular space. Their large, complex structure, stabilized by disulfide bonds, is not suited for the reducing environment inside a cell, where they often misfold and become non-functional. This has created a significant blind spot, preventing researchers from using these highly specific binders to study the vast and complex world of intracellular proteins in their native context.
Intrabodies, or intracellular antibodies, were conceived to overcome this limitation, but their development has been notoriously difficult, plagued by low success rates and unpredictable performance. The new AI-powered framework directly confronts this issue by re-engineering the antibody structure for intracellular stability from the ground up. By combining predictive modeling with intelligent sequence redesign, this approach provides a systematic and reliable solution, transforming a process once defined by trial and error into a predictable engineering discipline. This technological shift is pivotal for unlocking new avenues in both fundamental research and therapeutic innovation.
Anatomy of the AI-Powered Design Pipeline
AI-Powered Protein Structure Prediction and Analysis
The foundation of the AI-driven pipeline rests on its ability to accurately predict the three-dimensional structure of a given antibody. Sophisticated deep learning models can generate a high-resolution structural blueprint from an amino acid sequence alone, a task that once required laborious and time-consuming experimental methods. This initial step is crucial, as it provides the essential spatial information needed to identify which parts of the antibody are critical for binding and which can be modified.
With a detailed 3D model in hand, the system can analyze the protein’s biophysical properties, identifying regions prone to instability or aggregation within the intracellular environment. This predictive analysis allows the pipeline to pinpoint vulnerabilities in the antibody’s framework—the structural scaffold that supports the antigen-binding loops. It is this precise, structure-based understanding that enables the subsequent redesign phase to be targeted and effective, rather than a speculative guessing game.
Intelligent Sequence Redesign for Intracellular Stability
The core innovation of the AI framework lies in its capacity to intelligently redesign the antibody sequence to promote proper folding and solubility inside the cell. Unlike traditional methods that might involve random mutations, the AI algorithm systematically suggests specific amino acid substitutions in the framework regions. These changes are carefully selected to enhance stability without altering the conformation of the complementarity-determining regions (CDRs), which are responsible for recognizing and binding the target antigen.
This process is a delicate balancing act, as the goal is to create a robust and functional molecule that retains its original specificity. The AI models evaluate thousands of potential mutations, predicting how each one will affect the protein’s overall stability and folding dynamics. By optimizing the antibody scaffold for the intracellular environment, the pipeline effectively “domesticates” the molecule, enabling it to function reliably where it previously could not.
Rapid Validation via Live-Cell Screening
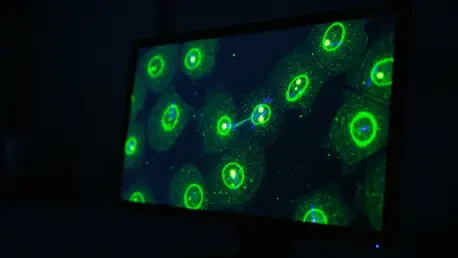
The final, and equally critical, component of the pipeline is the high-throughput validation of the newly designed intrabodies within living cells. This empirical testing phase closes the design-build-test loop, providing immediate feedback on the performance of the AI-generated candidates. Engineered intrabodies are introduced into cells and tagged with fluorescent markers, allowing researchers to directly observe their expression, stability, and localization.
This live-cell screening approach offers a significant advantage over in vitro methods, as it confirms functionality in the exact context where the intrabody is intended to be used. By monitoring fluorescence levels and binding patterns, scientists can quickly determine whether the redesigned intrabody is stable, soluble, and specific to its target. This rapid validation cycle dramatically accelerates the development process, enabling the efficient identification of successful candidates from a pool of designs.
Performance Breakthroughs and Validation
Revitalizing Failed Candidates into Functional Intrabodies
One of the most compelling demonstrations of the AI pipeline’s power is its ability to salvage antibody sequences that had previously failed to function as intrabodies. In recent validation studies, researchers successfully converted 19 out of 26 existing antibody sequences into effective intracellular tools. Remarkably, 18 of these had been written off as non-starters using conventional development approaches, highlighting the transformative potential of this new methodology.
This success rate not only validates the AI-driven design principles but also suggests that a vast repository of existing antibodies, previously deemed unsuitable for intracellular work, can now be revisited. The ability to “resurrect” these failed candidates opens up a treasure trove of well-characterized binders for new applications, saving immense time and resources that would otherwise be spent on discovering novel antibodies from scratch.
Real-Time Monitoring of Dynamic Cellular Processes
The practical utility of these AI-designed intrabodies has been powerfully demonstrated in their application to monitor dynamic biological events in real time. A key success story involves the creation of intrabodies that target specific histone modifications—chemical tags on proteins that package DNA and play a crucial role in regulating gene activity. These new probes allow scientists to visualize the ebb and flow of these epigenetic markers as cells respond to stimuli.
By linking the intrabodies to fluorescent proteins, researchers can directly observe changes in gene expression patterns inside a single living cell, a feat that was previously difficult to achieve with such specificity and temporal resolution. The fluorescence intensity of the intrabody directly correlates with the abundance of its target, providing a dynamic readout of cellular state. This capability offers an unprecedented window into the complex regulatory networks governing cell function, health, and disease.
Real-World Applications and Scientific Impact
Next-Generation Live-Cell Imaging and Diagnostics
The AI-designed intrabodies are poised to become next-generation probes for advanced live-cell imaging and diagnostics. Their high specificity and stability make them superior to many existing tools, enabling researchers to label and track intracellular molecules with exceptional clarity. In fluorescence microscopy, these intrabodies can illuminate the precise location and movement of proteins, offering deeper insights into cellular organization and signaling pathways.
Beyond basic research, this technology holds promise for the development of novel cellular diagnostics. By designing intrabodies that recognize disease-specific biomarkers inside patient cells, it may become possible to create highly sensitive tests for early disease detection or to monitor therapeutic responses at the single-cell level. This could lead to more precise and personalized diagnostic strategies for a variety of conditions, including cancer and neurodegenerative disorders.
A New Frontier for Intracellular Therapeutics
Perhaps the most exciting long-term prospect for AI-driven intrabody design is the development of novel intracellular therapeutics. Many diseases are caused by malfunctioning proteins located inside cells, which have historically been considered “undruggable” by conventional antibody therapies. By engineering intrabodies that can enter cells and specifically neutralize these disease-causing targets, a new class of powerful medicines could be created.
This approach could open up therapeutic avenues for a wide range of illnesses, from viral infections, where intrabodies could block viral replication, to cancers driven by aberrant internal proteins. While significant challenges remain in terms of delivery and clinical translation, the ability to reliably design functional intrabodies is a critical first step toward realizing this vision. It represents a new frontier in drug discovery, moving beyond the cell surface to tackle disease at its source.
Current Challenges and Technical Hurdles
Ensuring Target Specificity and Minimizing Off-Target Effects
Despite the pipeline’s successes, ensuring absolute target specificity remains a critical challenge. The intracellular environment is incredibly crowded, containing thousands of different proteins. An intrabody must bind exclusively to its intended target without engaging in off-target interactions that could generate misleading data in research applications or cause toxic side effects in therapeutic contexts.
While the AI preserves the original antigen-binding sites, subtle conformational changes or non-specific stickiness can still lead to unintended binding events. Mitigating this risk requires a combination of sophisticated computational prediction of potential off-targets and rigorous experimental validation. Ongoing efforts are focused on refining the AI algorithms to better predict and minimize these off-target effects, ensuring that each designed intrabody is not only stable but also exceptionally precise.
Scalability, Accessibility, and Computational Costs
For AI-driven intrabody design to become a mainstream tool, practical hurdles related to scalability and accessibility must be addressed. The deep learning models and molecular dynamics simulations at the heart of the pipeline require substantial computational resources, which can be a significant barrier for many academic labs and smaller biotech companies. The high costs associated with this computing power could limit the widespread adoption of the technology.
Furthermore, democratizing this technology requires creating user-friendly platforms and workflows that do not demand deep expertise in computational biology. Efforts to streamline the pipeline, optimize algorithms for efficiency, and develop cloud-based platforms will be essential in making these powerful design tools available to the broader scientific community. Bridging the gap between cutting-edge development and practical application is key to unlocking the full potential of this innovation.
Future Directions and Long-Term Vision
Unlocking Vast Antibody Databases for Intracellular Use
Looking ahead, a major opportunity lies in applying the AI pipeline to the vast, publicly available databases of antibody sequences that have been curated over decades. These libraries contain sequences for antibodies targeting a massive array of antigens, but most have never been tested for intracellular function. A systematic, large-scale effort to process these sequences through the AI design framework could rapidly expand the toolkit of available intrabodies.
Such an initiative could generate a comprehensive catalog of functional intracellular probes targeting thousands of different human proteins. This would provide the research community with an invaluable, off-the-shelf resource for studying virtually any cellular process. Transforming these existing data repositories into a functional toolkit represents a powerful and efficient path toward mapping the human intracellular proteome in living cells.
Accelerating Personalized Medicine and Drug Discovery
The long-term vision for this technology is its integration into the paradigm of personalized medicine. As our ability to identify patient-specific or disease-specific protein targets grows, the AI pipeline could enable the rapid creation of custom intrabodies tailored to an individual’s unique molecular profile. This could lead to hyper-personalized therapeutics that are designed to be maximally effective and minimally toxic for a specific person.
In the broader context of drug discovery, this platform could dramatically accelerate the development of new biologic drugs. By replacing the slow, trial-and-error process of intrabody engineering with a fast, predictable, and automated design cycle, the timeline for moving from target identification to a viable therapeutic candidate could be significantly shortened. This acceleration promises to bring novel treatments to patients faster and more efficiently than ever before.
Conclusion: A Paradigm Shift in Cellular Investigation
The development of an AI-driven pipeline for intrabody design marked a genuine paradigm shift in our ability to probe and manipulate the inner workings of the cell. By successfully overcoming the long-standing challenges of intracellular protein stability, this technology transformed what was once an artisanal and unreliable process into a systematic and predictable engineering discipline. The demonstrated ability to revitalize previously failed antibodies and create high-fidelity probes for real-time cellular monitoring underscored its immediate impact on biological research.
This breakthrough has paved the way for profound advancements across multiple fields. In diagnostics, it promised a new generation of cell-based assays with unprecedented specificity, while in therapeutics, it opened the door to targeting intracellular disease-causing molecules that were previously considered inaccessible. Although challenges related to specificity and scalability remained, the fusion of artificial intelligence and protein engineering had fundamentally expanded the toolkit for cellular investigation, setting the stage for future discoveries and innovations in medicine.