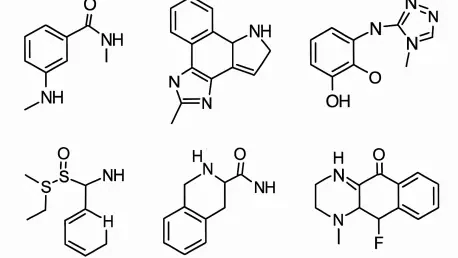
In the intricate landscape of drug discovery, pinpointing effective inhibitors for Protein Tyrosine Phosphatase 1B (PTP1B) stands as a monumental challenge with profound implications for treating metabolic disorders such as diabetes and obesity. PTP1B is a key player in regulating insulin signaling pathways, positioning it as a vital therapeutic target. Yet, the journey to predict the activity of potential inhibitors has been fraught with obstacles, largely due to the striking differences between natural products derived from biological sources and synthetic compounds crafted in laboratories. A recent study published in Molecular Diversity by Wang, Sun, Ren, and colleagues unveils a pioneering solution through a transfer learning framework, setting a new benchmark in predictive accuracy. This innovative approach not only tackles long-standing hurdles in pharmacological research but also opens doors to accelerated drug development, promising significant advancements for both researchers and patients grappling with metabolic conditions.
Innovations in Predictive Modeling
Overcoming Structural Diversity
The core challenge in predicting PTP1B inhibitor activity lies in the vast structural diversity between natural and synthetic compounds, a disparity that traditional models often fail to navigate effectively. Natural products, shaped by evolutionary processes, frequently exhibit intricate molecular architectures that interact with biological targets in unique ways. Synthetic compounds, by contrast, are engineered with specific chemical traits, often lacking the nuanced binding capabilities of their natural counterparts. This dichotomy has historically led to inconsistent predictions, as conventional modeling techniques struggle to adapt to such varied chemical spaces. The transfer learning framework introduced in the study offers a transformative solution by drawing on knowledge from broader, related datasets. By integrating insights from diverse chemical domains, this method enhances the model’s ability to account for structural differences, resulting in more accurate predictions of inhibitor efficacy across both compound types.
Another critical aspect of this approach is how it bridges the gap between disparate chemical categories, ensuring that the predictive model remains versatile and reliable. Transfer learning leverages pre-existing data from related tasks to bolster performance in areas with limited specific information, such as PTP1B inhibition. This strategy allows the framework to capture subtle molecular interactions that might be missed by models trained solely on narrow datasets. The study demonstrates that this adaptability is particularly beneficial when dealing with the complex binding mechanisms of natural products versus the more targeted designs of synthetic molecules. As a result, researchers gain a more comprehensive tool for identifying potential inhibitors, reducing the guesswork in early-stage drug discovery. This advancement marks a significant shift from traditional methodologies, offering a robust foundation for tackling one of the most persistent challenges in medicinal chemistry.
Battling Data Scarcity and Overfitting
Data scarcity remains a formidable barrier in pharmacological research, especially for specialized targets like PTP1B, where comprehensive datasets are often unavailable. Small data pools can severely limit the effectiveness of predictive models, making it difficult to draw meaningful conclusions about a compound’s potential as an inhibitor. The transfer learning framework addresses this issue head-on by incorporating knowledge from larger, related datasets, thereby enriching the training process. This approach ensures that the model is exposed to a wider array of chemical and biological information, enhancing its capacity to make accurate predictions even when specific PTP1B data is sparse. Such an innovation is crucial for advancing drug discovery, as it allows scientists to work with limited resources while still achieving reliable outcomes, paving the way for more efficient research pipelines.
Equally important is the framework’s ability to mitigate the risk of overfitting, a common pitfall when models are trained on small datasets. Overfitting occurs when a model becomes overly tailored to its training data, losing its ability to generalize to new, unseen compounds. This can lead to misleading predictions and hinder progress in identifying viable inhibitors. By utilizing transfer learning, the study’s model avoids this trap, maintaining robustness across diverse chemical spaces. The methodology fine-tunes its predictions by balancing learned patterns from broad datasets with the specifics of PTP1B inhibition, ensuring consistency and reliability. This dual focus on combating data scarcity and preventing overfitting represents a substantial leap forward, equipping researchers with a tool that not only addresses current limitations but also sets a precedent for handling similar challenges in other areas of drug development.
Practical Applications and Accessibility
Empowering Researchers with a Web Platform
One of the most impactful outcomes of this research is the creation of a user-friendly web platform that democratizes access to the predictive model for PTP1B inhibitors. This online tool allows scientists from various backgrounds to input compound structures and receive detailed predictions about their potential efficacy. Such accessibility is a game-changer, particularly for researchers in academia or smaller institutions who may lack the computational infrastructure or expertise to develop similar models independently. By removing these barriers, the platform ensures that cutting-edge technology is within reach of a global audience, fostering an inclusive environment for drug discovery. The ease of use and immediate feedback provided by the tool can significantly accelerate the initial screening process, helping to identify promising candidates for further experimental validation.
Beyond individual access, the web platform serves as a catalyst for streamlining research workflows across the scientific community. Researchers can quickly test hypotheses about potential inhibitors without the need for extensive preliminary experiments, saving both time and resources. This efficiency is particularly vital in the fast-paced field of pharmacology, where delays can impede the development of life-saving treatments. Additionally, the platform’s design prioritizes user experience, ensuring that even those with minimal technical background can navigate it effectively. By providing a centralized resource for predictive analysis, the tool not only enhances productivity but also encourages the sharing of insights and data among peers, amplifying its impact. This practical implementation underscores the study’s commitment to translating complex research into tangible benefits for the broader scientific landscape.
Expanding Collaborative Horizons
Collaboration lies at the heart of this innovation, with the web platform acting as a bridge connecting diverse sectors of the scientific community, including academia, biotechnology, and the pharmaceutical industry. By making the predictive model widely available, the platform fosters a spirit of shared progress, enabling interdisciplinary teams to work together toward common goals. Researchers can exchange findings, validate predictions, and build on each other’s work, creating a synergistic effect that accelerates drug discovery. This collaborative approach is essential for tackling the multifaceted challenges of developing effective PTP1B inhibitors, as it combines expertise from computational science, pharmacology, and medicinal chemistry into a unified effort. The result is a more dynamic research ecosystem that thrives on collective knowledge and innovation.
Furthermore, the platform’s role in promoting collaboration extends to its potential for global impact, breaking down geographical and institutional barriers. Scientists from different regions and backgrounds can contribute to and benefit from the predictive tool, ensuring that advancements in PTP1B inhibitor research are not confined to well-resourced hubs. This inclusivity can lead to a more diverse pool of compounds being tested, increasing the likelihood of discovering novel inhibitors with unique properties. The emphasis on shared access also aligns with broader trends in scientific research, where open tools and data-sharing initiatives are becoming integral to progress. By championing collaboration, the study not only enhances the immediate application of its framework but also sets a model for how technology can unite efforts in solving complex health challenges, ultimately benefiting a wider population.
Future Prospects and Broader Impact
Extending the Framework’s Reach
Looking to the horizon, the researchers behind this study envision expanding the transfer learning framework beyond PTP1B to encompass other therapeutic targets, potentially revolutionizing predictive modeling in medicinal chemistry. This methodology, with its ability to adapt to limited data and diverse chemical spaces, holds promise for addressing a range of biological challenges where traditional approaches fall short. Targets related to cancer, neurodegenerative diseases, or other metabolic conditions could benefit from similar strategies, leveraging shared knowledge from related datasets to improve prediction accuracy. Such an expansion would mark a significant shift in how drug discovery is conducted, positioning machine learning as a cornerstone of future research. The scalability of this framework suggests a transformative impact, offering a blueprint for tackling complex problems across various domains of pharmacology.
Additionally, the continuous refinement of this approach is a priority, with plans to integrate more data and enhance model capabilities over time. By incorporating feedback from users of the web platform and conducting further validation studies, the framework can evolve to address emerging needs in drug development. This iterative process ensures that the technology remains relevant and effective as new therapeutic challenges arise. The potential to apply transfer learning to uncharted areas of medicinal chemistry also opens avenues for innovation, encouraging researchers to explore novel targets and mechanisms of action. This forward-thinking perspective not only amplifies the study’s current contributions but also establishes a foundation for long-term advancements, signaling a new era where computational tools play an integral role in navigating the complexities of biological systems.
Enhancing Patient Outcomes
The ultimate significance of this research lies in its potential to improve health outcomes for patients, particularly those battling metabolic disorders like diabetes and obesity. With more accurate predictive tools for PTP1B inhibitors, the development of targeted therapies can be expedited, reducing the time it takes to bring effective treatments to market. This acceleration is critical for addressing urgent medical needs, offering hope to millions who rely on innovative solutions for better quality of life. The ability to identify promising inhibitors early in the research process also means that resources can be allocated more efficiently, focusing on compounds with the highest likelihood of success. For patients, this translates to faster access to personalized treatments that address the root causes of their conditions.
Moreover, the broader implications of this study extend to the promise of precision medicine, where therapies are tailored to individual needs based on robust predictive data. The transfer learning framework, by enhancing the reliability of early-stage drug screening, supports the creation of treatments that are more effective and less prone to side effects. This patient-centered focus is a driving force behind the research, aligning with the overarching goal of improving health care through scientific innovation. As the framework’s applications grow to include other therapeutic areas, its impact on patient care could become even more profound, heralding a future where computational advancements directly contribute to tangible health benefits. This vision of enhanced outcomes reflects the study’s lasting contribution to both science and society, emphasizing the power of technology to transform lives.